Quantum hardware has made significant advances in the last few years, but finding compelling applications with practical quantum advantage has been challenging. In today’s article, we argue that real applications will be a hybrid combination of quantum and classical computation, and that the quantum and classical algorithms for these computations need to be co-designed with each other. Moreover, we explore the novel application space of finding biomarkers in multimodal cancer data, where even the classical algorithms are still under development. We propose that quantum-classical algorithmic co-design can be applied from the beginning of application development, resulting in quantum-inspired and quantum-compatible classical algorithms that are already showing promising results.
Real quantum applications will be hybrid and co-designed with domain experts
Quantum computers are domain-specific accelerators. It is expected that they will only outperform classical algorithms for a subset of computational tasks. Discovering which tasks yield such a speedup is an ongoing field of research, but some notable examples include quantum algorithms for factoring large integers and computing discrete logarithms, sampling from complicated probability distributions, as well as simulating the time dynamics and computing expectation values of entangled quantum states. Recent research has also explored quantum algorithms for machine learning tasks, leading to provable memory separations between quantum and classical models for sequential learning tasks [Gao+22, Anschuetz+23]. Additionally, the development of heuristic algorithms for optimization problems has shown promising results. A shared aspect amongst all of these algorithms is that the quantum portion of the computation is always embedded as a subroutine within a larger application, running on classical hardware, aimed at solving a problem of practical interest.\
Quantum computers are domain-specific accelerators. It is expected that they will only outperform classical algorithms for a subset of computational tasks. Discovering which tasks yield such a speedup is an ongoing field of research, but some notable examples include tasks such as factoring large integers and computing discrete logarithms, sampling from complicated probability distributions, as well as simulating the time dynamics and computing expectation values of entangled quantum states. In general, these computational tasks appear as subroutines embedded within some larger application, running on classical hardware, aimed at solving a problem of practical interest.
These are all examples of hybrid applications wherein classical and quantum co-processors share different aspects of a user workload. Determining the optimal partitioning and balancing of that workload is an interesting open problem, one that is constantly changing as hardware capabilities continue to improve, and a prime target for co-design approaches as discussed below. In addition to the notion of hybrid workload-sharing applications, quantum applications are also hybrid in the sense that they often require classical computation to support the quantum computation in various ways. For example, error correction, error mitigation, and the low-level control logic for the quantum computer are all techniques that involve classical co-processing to support the proper functioning of the quantum processor.
Due to limited quantum computational resources (e.g., limited qubit count and program depth) and the overhead of classical co-processing, practically useful quantum applications will employ the quantum processor as a domain-specific accelerator. The goal of the co-design approach to hybrid algorithm design is to first identify the computational task within the application that is most amenable to quantum speedup, and then design the algorithm in such a way that respects the limitations of the quantum hardware and minimizes the classical co-processing overhead, while still retaining (at least some of) the original quantum speedup.
Designing effective quantum applications that retain their theoretically achievable speedups in practice is a challenging task. This is evidenced by recent work which showed that the quadratic speedup of Grover’s algorithm would likely be lost for practical implementations taking into account the overhead induced by error correction. Similarly, the quantum speedup offered by other quantum algorithms, such as the HHL algorithm for solving linear systems, may be lost when considering the overhead needed to prepare input states in a fully end-to-end implementation. We can begin to overcome these challenges by closely collaborating with domain experts to design end-to-end hybrid applications that apply quantum accelerators to specific computational bottlenecks and account for overhead costs with the aim of yielding practical quantum speedups.
Hybrid applications for multi-modal cancer data analysis
Taking a broad view, there are many potential applications of quantum algorithms to cancer data analysis. Using a quantum computer to efficiently simulate the time dynamics of molecular systems may be used to improve in silico modeling of small molecules and proteins for new therapies [Alevras+, Kao+]. Quantum algorithms for optimization and machine learning may be used to more efficiently model the correlations within cancer data sets with applications to dimensionality reduction, clustering, and predicting treatment outcomes. Furthermore, quantum sensing technologies may provide even more detailed imaging, metabolic monitoring, and therapy response characterization at a cellular level.
One particularly promising example for quantum-classical co-design is the discovery of biomarkers in multimodal cancer data, a research effort within Wellcome-Leap’s Quantum for Bio program. Biomarkers are biological indicators that can be used to detect the presence or progression of a disease, such as cancer. However, identifying these biomarkers in large, complex datasets is a daunting task that requires sophisticated computational techniques. Even the classical algorithms for this purpose are still under development, presenting an opportunity for quantum-classical co-design to make a significant impact.
Focus: accelerating feature selection for biomarker discovery
Analyzing multimodal cancer data presents two significant computational challenges. One major difficulty lies in accurately modeling the exponentially many ways in which the features may interact within or between biologically related data modes. Additionally, compared to domains like natural language processing, cancer data is relatively scarce – while each patient data point may be extremely high-dimensional, spanning gigapixel images and thousands of genomic features, the overall number of patients is typically much smaller than the number of features due to the expense and time required for data collection, making it challenging for machine learning models to robustly analyze the data without overfitting. This is an interesting scenario in which increased solution quality is more the focus rather than a quantum speed advantage; although these are not entirely orthogonal since classical computers can simulate small quantum algorithms with an exponential speed penalty.

The figure above outlines a machine learning pipeline that may be used for biomarker discovery. We will focus on the second step which selects a subset of features from the raw data which will then be fed into a downstream machine learning model. For data scarce ML applications such as this, dimensionality reduction is a key part of the pipeline in order to ensure robust generalizable learning. While other methods, such as principal component analysis or autoencoders, can be used to similarly reduce the total number of features, they also transform the original features in a way that may reduce interpretability. Instead, we focus on filter methods for feature selection that use a combinatorial optimization framing of the problem and select a strict subset of the original raw features to participate in the downstream learning problem, thereby preserving interpretability.
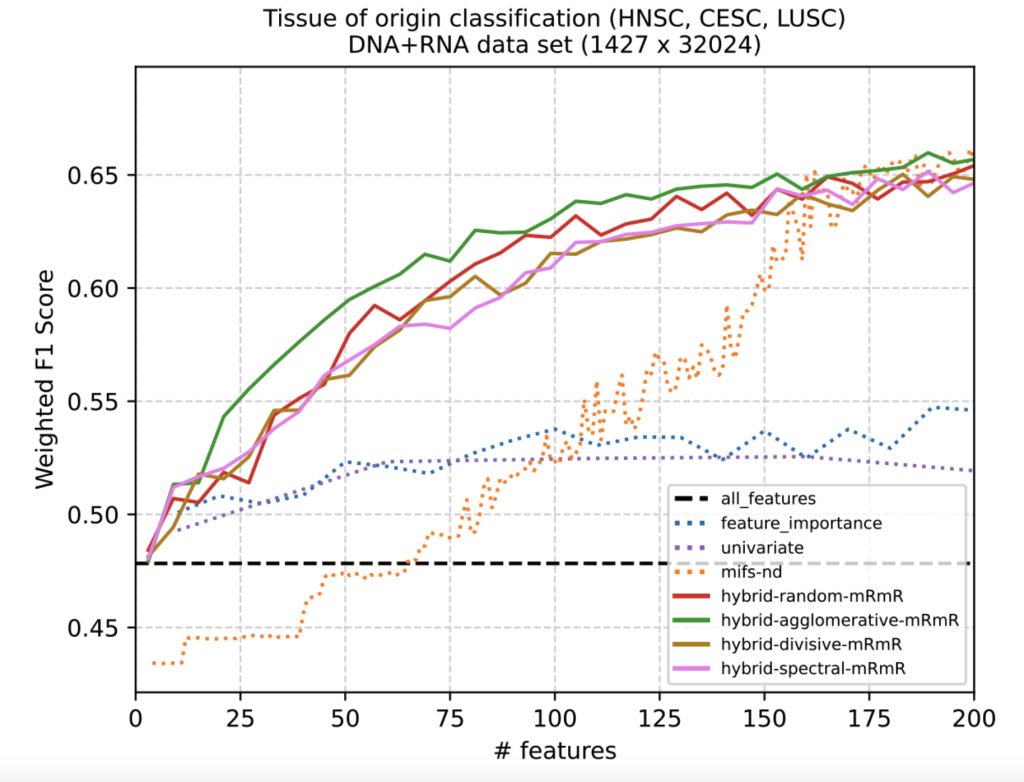
The figure above shows some early results on the performance of hybrid quantum-classical feature selection algorithms compared to classical baseline algorithms for a multi-modal DNA and RNA data set. The hybrid algorithms (solid lines) use quantum optimization heuristics and are able to select smaller feature sets that yield greater classification accuracy than feature sets of equal sized produced by classical algorithms (dashed lines). What remains to be seen is how these feature sets will interact with downstream classical and quantum machine learning approaches for predicting cancer tissue of origin or treatment outcomes. We expect an iterative co-design process to refine the interaction between quantum and classical algorithms in the data pipeline, as well as the types of cancer and outcomes as a problem focus.
A future of quantum accelerators
The collaboration between quantum computing experts and cancer researchers in feature selection for multimodal data serves as a compelling example of the potential for domain-specific quantum accelerators. As we look to the future of computing, it is becoming increasingly clear that heterogeneous systems, composed of multiple accelerators such as CPUs, GPUs, and QPUs, will play a crucial role in achieving continued performance gains. As the capabilities of quantum processors continue to grow, the balance of computational workloads across these accelerators is likely to shift. This presents an exciting architectural and algorithm design challenge: designing tools and applications that seamlessly optimize workload allocation, matching computational kernels to the most suitable accelerators. By collaborating with domain experts and designing systems in this codesign fashion, we can effectively accelerate a wide range of meaningful applications and continue to realize computational performance improvements.
About the Authors:
Teague Tomesh is a Manager of Quantum Software Engineering at Infleqtion. He is a Principal Investigator of projects funded by Wellcome Leap’s Q4Bio program, DARPA IMPAQT, and DOE EXPRESS.
Sid Ramesh is a Resident Physician-Scientist at the University of Chicago Medicine, practicing internal medicine and medical oncology. His research focuses on applying artificial intelligence and deep learning techniques to cancer diagnosis and treatment, particularly in areas such as predicting cancer progression in oral premalignant lesions and outcomes in neuroblastoma patients.
Colin Campbell is a Quantum Applications Engineer at Infleqtion. He is an expert in quantum walks, machine learning, and optimization algorithms with deep experience co-designing hybrid algorithms for various domains including multimodal cancer and quantum chemistry applications.
Alex Pearson is a practicing medical oncologist, Director of Head/Neck Cancer, Director of Heme/Onc Data Sciences, and Associate Professor of Medicine at UChicago. Dr. Pearson is trained as a PhD statistician as well as an MD physician, and maintains joint appointments at Argonne National Laboratory, UChicago Comprehensive Cancer Center, and the Section of Biomedical Data Science.
Samantha Riesenfeld is an Assistant Professor in the UChicago Pritzker School of Molecular Engineering and in the Department of Medicine, Section of Genetic Medicine with additional affiliations in interdepartmental research centers including the Committee on Immunology, for which she co-chairs the Computational and Systems Immunology Subcommittee, the Institute for Biophysical Dynamics, the Data Science Institute, and the Comprehensive Cancer Center.
Fred Chong is the Seymour Goodman Professor of Computer Architecture at the University of Chicago and the Chief Scientist for Quantum Software at Infleqtion. He is the Lead Principal Investigator of the (Enabling Practical-scale Quantum Computation), an NSF Expedition in Computing, as well as the Lead PI of a Wellcome-Leap Q4Bio project. He is also an advisor to Quantum Circuits, Inc.
Disclaimer: These posts are written by individual contributors to share their thoughts on the Computer Architecture Today blog for the benefit of the community. Any views or opinions represented in this blog are personal, belong solely to the blog author and do not represent those of ACM SIGARCH or its parent organization, ACM.


