More than a decade ago, the pioneering computer scientist Jim Gray envisioned a fourth paradigm of scientific discovery that uses computers to solve data-intensive scientific problems. Today, scientific discovery, whether observational, in-silico or experimental, requires sifting through and analyzing complex, large datasets. For example, plasma simulation simulates billions of particles in a single run, but analyzing the results requires sifting through a single frame (a multi-dimensional array) that is more than 50 TB big—and that is for only one timestep of a much longer simulation. Similarly, modern observation instruments also produce large datasets: a two-photon imaging of a mouse brain yields up to 100 GB of spatiotemporal data per hour and electrocorticography (ECoG) recordings yield 280 GB per hour.
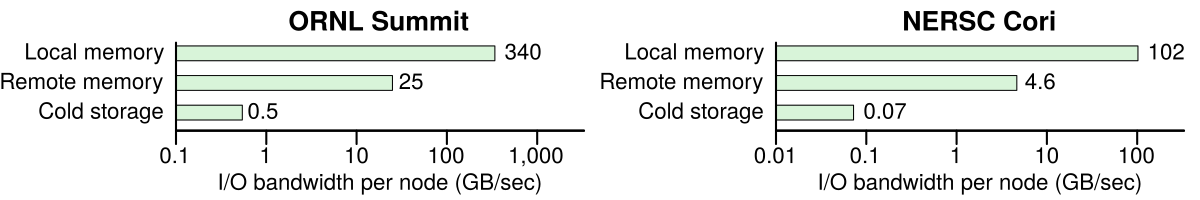
Many scientific applications are increasingly becoming I/O bound, as the bandwidth to cold storage is 50× – 1,500× less than the bandwidth to memory. Per-node bandwidth is shown for Summit and Cori, #1 and #13 computers in TOP500, respectively. Data from Vazhkudai et al. and NERSC.
Although scientific computing continuously shatters records for floating point performance, the I/O capabilities of scientific computers significantly lag on-premise datacenters and the cloud. As shown in the figures above, memory is 1,000× slower than cold storage in the fastest scientific computers today. Making matters worse, the reported I/O performance assumes large sequential I/Os, although many applications do not exhibit this I/O pattern. For example, classification in scientific applications is often performed on millions of KB-sized objects, which poorly utilizes the file system. Given the exponential growth trend in data volumes, the bottleneck for many scientific applications is no longer floating point operations per second (FLOPS) but I/O operations per second (IOPS).
The computational side of a scientific computer is undergoing a rapid transformation to embrace array-centric computing all the way from applications to the hardware. However, the POSIX I/O interface to cold data remains largely agnostic to the I/O optimization opportunities that array-centric computing presents. This simplistic view of I/O for array-centric analyses is challenged by the dramatic changes in hardware and the diverse application and analytics needs in today’s large-scale computers: On the hardware front, new storage devices such as SSDs and NVMe not only are much faster than traditional devices, but also provide a different performance profile between sequential and random I/Os. On the application front, scientific applications have become much more versatile in both access patterns and requirements for data collection, analysis, inference and curation. Both call for more flexible and efficient I/O abstractions for manipulating scientific data.
The rising prominence of file format libraries
Traditionally, scientific applications performed I/O in an application-specific manner that was hard to optimize systematically: storage structures were tailored to specific problems, there was no agreement on how data are represented, and data access methods would not be reused across projects. This lack of convergence has been attributed to many external factors, such as the significant upfront effort investment to use new software, the lack of computer science expertise, an underestimation of the difficulty of fundamental data management problems, and the limited ability of scientists to provide long-term support for their own code. As a consequence, scientists optimize I/O by following a simple “place data sequentially” mantra: data that are going to be accessed together are placed adjacently, while the system ensures that accesses to sequential data are fast through request batching and prefetching. Alas, this I/O optimization model is overly simplistic given the increasing heterogeneity of the storage stack that includes burst buffers, all-flash storage, and non-volatile memory.
The rapid adoption of new storage media has shifted the perception on using third-party I/O libraries for data storage and access. A 2014 user survey of the NERSC scientific facility shows that the HDF5 and NetCDF file format libraries are among the most widely used libraries, and have as at least as many users as the well-known numerical libraries BLAS, ScaLAPACK, MKL and fftw. In 2018, 17 out of the 22 projects in the DoE Exascale Computing Project (ECP) portfolio wanted to use the HDF5 file format. These file format libraries present a semantically richer, array-centric data access interface to scientific applications. Using such file format libraries allows applications to organize datasets in hierarchies, associate rich metadata with each dataset, retrieve subsets of an array, perform strided accesses, transform data, make value-based accesses, and automatically compress sparse datasets to save space—all while remaining oblivious to where and how the data is physically stored.
Many research efforts are developing data management functionality for established file formats. They largely explore three complementary avenues: (1) developing mechanisms to support user-defined functionality, (2) developing connectors and (3) automatically sharding, placing and migrating data between storage devices. Focusing on the HDF5 format, the ExaHDF5 project seeks to extend the HDF5 file format library with user-defined functionality. ExaHDF5 has developed the virtual object layer (VOL) feature which permits system builders to intercept and respond to HDF5 I/O calls. ArrayBridge is a connector that allows SciDB to directly query HDF5 data without the onerous loading step and produce massive HDF5 datasets using parallel I/O from within SciDB. Automatic migration between data storage targets has been proposed for HDF5 with the Data Elevator that transparently moves datasets to different storage locations, such as node-local persistent memory, burst buffers, flash, disks and tape-based archival storage.
Rethinking data storage
Storage providers are quickly innovating to reduce latency and significantly improve throughput. I/O optimizations such as coalescing, buffering, prefetching and aggregation optimize accesses to block-based devices, but incur unnecessary overhead for next-generation storage devices. New storage technologies will thus reach their full potential only if they reduce I/O stack overheads with direct user-mode access to hardware. Today applications can directly interact with new storage hardware by using libraries such as the persistent memory development kit (PMDK) and the storage performance development kit (SPDK). In the meantime, storage interfaces such as NVMe are being extended to support datacenter-scale connectivity with NVMe over Fabrics (NVMe-OF), which ensures that the network itself will not be the bottleneck for solid state technologies. This motivates a reconsideration of the data storage architecture of datacenter-scale computers.
An idea that is seeing significant application traction is to transition away from block-based, POSIX-compliant file systems towards scalable, transactional object stores. The Intel Distributed Asynchronous Object Storage (DAOS) project is one such effort to reinvent the exascale storage stack. DAOS is an open source software-defined object store that provides high bandwidth, low latency and high I/O operations per second. DAOS aggregates multiple storage devices (targets) in pools, which are also the unit of redundancy. The main storage entity in DAOS is a container, which is an object address space inside a specific pool. Containers support different schemata, including a filesystem, a key/value store, a database table, an array and a graph. Each container is associated with metadata that describe the expected access pattern (read-only, read-mostly, read/write), the desired redundancy mechanism (replication or erasure code), and the desired striping policy. Proprietary object-store solutions, such as the WekaIO Matrix, are also strongly competing in the same space. In these new storage designs, POSIX is no longer the foundation of the data model. Instead, POSIX interfaces are built as library interfaces on top of the storage stack, like any other I/O middleware.
Many challenges remain in how applications interact with persistent storage. One point of significant friction has been how applications make writes durable. Block-based interfaces rely on page-level system calls, such as fsync and msync, however flushing an entire page is too coarse-grained for byte-addressable non-volatile storage. Flushing at a finer granularity is possible with user-space instructions such as CLFLUSH that avoids an expensive system call. However, cache line flushing is not a panacea because it evicts lines from the cache. This means that accessing a location immediately after flushing it will cause a cache miss, which doubles the cost of a store. A more elegant solution currently in the works is eADR, which extends the power failure protection domain to include the cache hierarchy, as shown in the figure below, but implementing eADR is more challenging for vendors.

eADR extends the power failure protection domain to the cache hierarchy and simplifies programming persistent memory, but requires additional support by vendors. Figure from this pmem.io blog post.
What does this mean for computer architects?
The evolution from a block-based to an object-based I/O stack presents new opportunities for computer architects. Simple transformations (such as converting a matrix from column-major order to row-major order) are time-consuming, which points to many opportunities for hardware offloading. An open question is how can hardware acceleration be leveraged for more complex I/O patterns, such as bitweaving, data-dependent record skipping, co-analysis and delta encoding. Looking further ahead, one opportunity is to design smart storage devices that expose application-level semantics (such as indexed access) through storage device interfaces.
About the author: Spyros Blanas is an Assistant Professor in the Department of Computer Science and Engineering at The Ohio State University. His research interest is high performance database systems.
Disclaimer: These posts are written by individual contributors to share their thoughts on the Computer Architecture Today blog for the benefit of the community. Any views or opinions represented in this blog are personal, belong solely to the blog author and do not represent those of ACM SIGARCH or its parent organization, ACM.


